ELIXIR supports the European Corona action plan and plays an important role in the development of the COVID-19 Data Portal. As the life-science data Research Infrastructure in Europe, ELIXIR is in a unique position to help increase the amount of publicly available Covid-related data and facilitate its processing, publication and reuse.
ELIXIR Belgium promotes and encourages the publication of all scientific data related to the Covid pandemic and provides the tools, know-how and brokering services for researchers to do so. Our first action is to support the submission of SARS-Cov-2 nucleotide sequences to public repositories.
To achieve this, we have collaboratively developed and compiled Galaxy tools and workflows necessary to clean, assemble and submit SARS-CoV-2 sequences to the European Nucleotide Archive (ENA). There are many advantages of using Galaxy including a graphical user interface, access to tools and workflows for pre-processing, downstream analysis and visualization of sequences (including SARS-CoV-2-specific ones, Maier et al., 2021). Galaxy also provides a platform for sharing of data and metadata, facilitating international collaboration, integration with other public resources and enabling publishing FAIR data and analysis workflows.
The tools
Human reads cleaning tool
In order to comply with Europe’s General Data Protection Regulation (GDPR), traces of human genetic information must be removed from the raw data before submitting it to ENA. We have wrapped Metagen-FastQC as a Galaxy tool for this purpose. More information on how to use this tool can be found on our Submission of SARS-Cov-2 raw reads page.
The ENA reads submission tool (command line)
Uploading raw reads to ENA can be done using the website, webin-CLI or programmatically through curl commands. Programmatic submissions are more convenient for bulk uploads, but require bioinformatic skills to generate the XML metadata files and to upload the data through ftp. To address this, ELIXIR Belgium together with the lab of Björn Grüning have developed a python command line interface (CLI) that:
- Makes submission easier for bioinformaticians
- Generates the required XML files out of easier-to-use tsv files or excel templates
- Takes care of the ftps uploading
- Validates the metadata before submission
- Can add, modify, cancel and release study, sample, experiment and run objects
The python package ena-upload-cli is published on pip and bioconda. For more information on how to use this tool please visit the github page of ena-upload-cli.
The Galaxy ENA reads submission tool
To make the process more user-friendly and allow most researchers without informatics experience to submit sequences to ENA without using command line, the tool was wrapped as a Galaxy tool. The ENA upload tool is part of the Intergalactic Utilities Commission (IUC), a curated collection of Galaxy tools. In this repository you can find all the information on how to install the tool yourself if you are administrator of a Galaxy instance. More information on how to use the tool itself can be found on our Submission of SARS-Cov-2 raw reads page.
COVID-19 genome analysis workflows
We have included the COVID-19 variant discovery and consensus building Galaxy workflows by Maier et al. (2021). They allow the analysis of Illumina WGS and amplicon as well as ONT amplicon data.
The Galaxy ENA consensus submission tool
A Galaxy wrapper of ENA Webin-CLI submission tool was made to submit SARS-CoV-2 consensus data to ENA. More information on how to use the tool itself can be found on our Submission of SARS-Cov-2 assemblies page.
All these tools and workflows are included in a custom Galaxy Docker container for ease of deployment. This guide describes processing and submission of SARS-CoV-2 sequence data using the tools and workflows in this container. A short introduction to Galaxy is recommended for users unfamiliar with the platform.
Overview of the submission process
The recommended workflow for SARS-CoV-2 sequence data process and submission using the Galaxy container is outlined in Figure 1. There are four main steps in this workflow:
- Remove human traces from SARS-CoV-2 sequences (Fig. 1a)
- Submit raw reads to ENA (Fig. 1b)
- Sequence analysis: variant detection and consensus building (Fig. 1c)
- Submit consensus sequences to ENA (Fig. 1d)
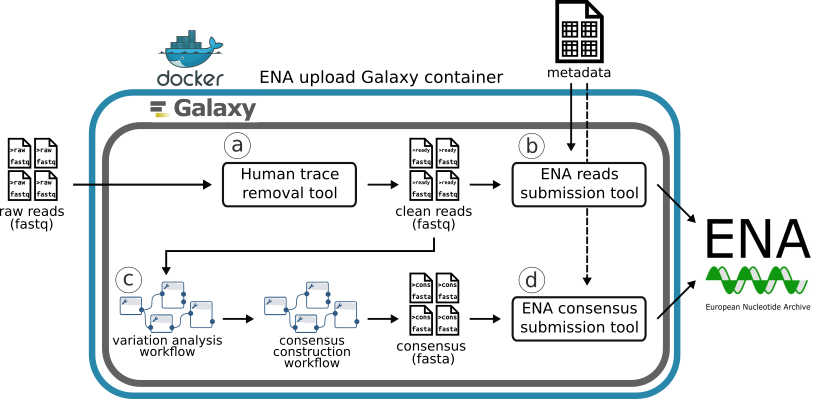
Figure 1. Recommended workflow for SARS-CoV-2 data process and submission.
We have divided the guide into two sections:
- Cleaning and submitting SARS-Cov-2 raw sequence reads to ENA and
- assembling and submitting consensus sequence of SARS-CoV-2 genomes to ENA
Support for Belgian researchers
We provide support for Belgian researchers to submit data as a service, through the brokering mechanism of ENA. To this end, we host the dedicated Galaxy instance covid19.useGalaxy.be, which contains all the tools and workflows discussed here. For more information or to request access credentials contact datasub at elixir-belgium.org.